log_link object provides a way to implement logarithmic link function that
maxlogL needs to perform estimation. See documentation for
maxlogL for further information on parameter estimation and implementation
of link objects.
Details
log_link is part of a family of generic functions with no input arguments that
defines and returns a list with details of the link function:
name: a character string with the name of the link function.g: implementation of the link function as a generic function inR.g_inv: implementation of the inverse link function as a generic function inR.
There is a way to add new mapping functions. The user must specify the details aforesaid.
See also
Other link functions:
NegInv_link(),
logit_link()
Examples
# One parameters of normal distribution mapped with logarithmic function
x <- rnorm(n = 10000, mean = 50, sd = 4)
theta_2 <- maxlogL( x = x, link = list(over = "sd",
fun = "log_link") )
summary(theta_2)
#> _______________________________________________________________
#> Optimization routine: nlminb
#> Standard Error calculation: Hessian from optim
#> _______________________________________________________________
#> AIC BIC
#> 56245.15 56259.57
#> _______________________________________________________________
#> Estimate Std. Error Z value Pr(>|z|)
#> mean 49.95824 0.04027 1240.5 <2e-16 ***
#> sd 4.02739 0.02848 141.4 <2e-16 ***
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#> _______________________________________________________________
#> Note: p-values valid under asymptotic normality of estimators
#> ---
# Link function name
fun <- log_link()$name
print(fun)
#> [1] "log"
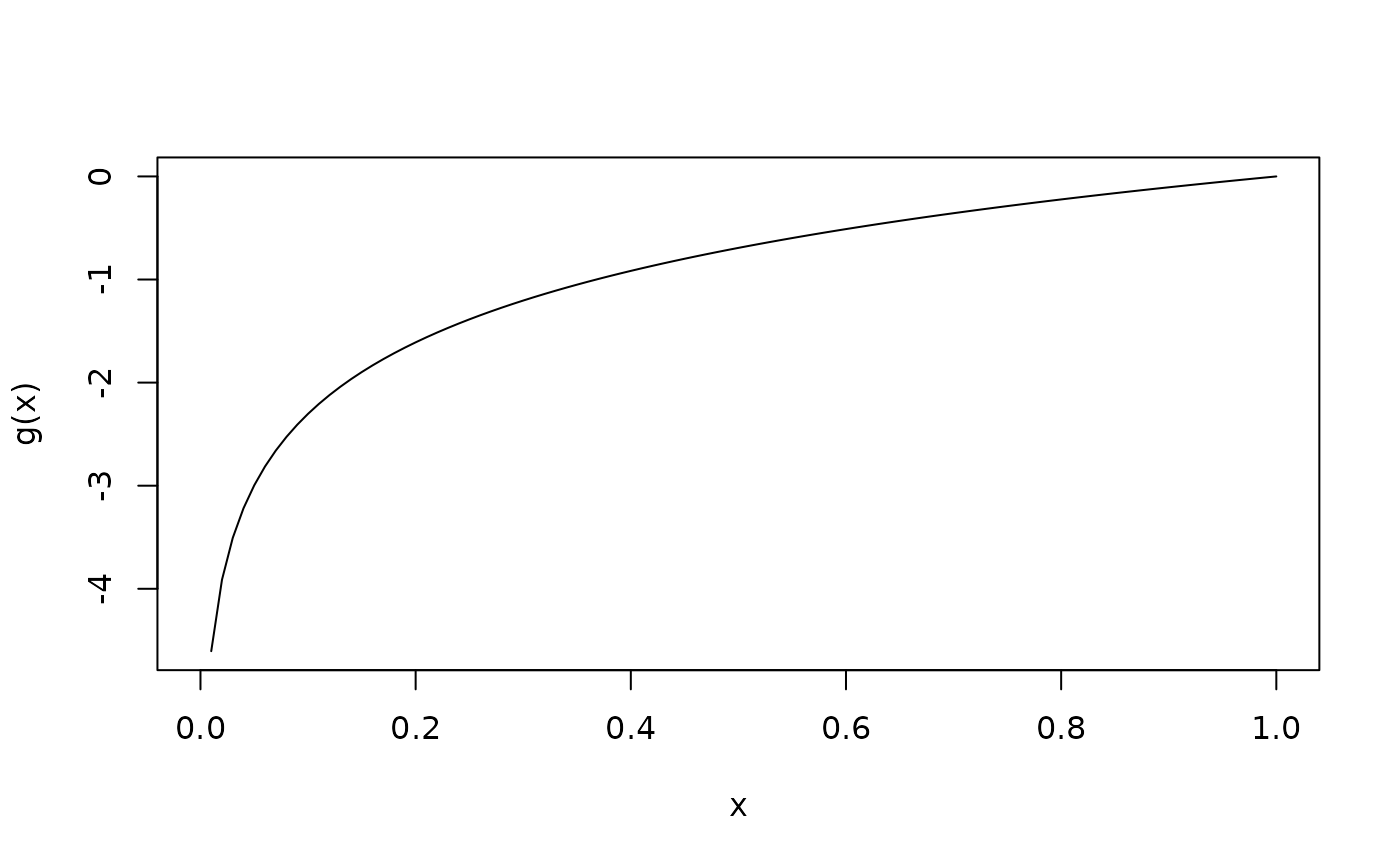
# Link function
g <- log_link()$g
curve(g(x), from = 0, to = 1)
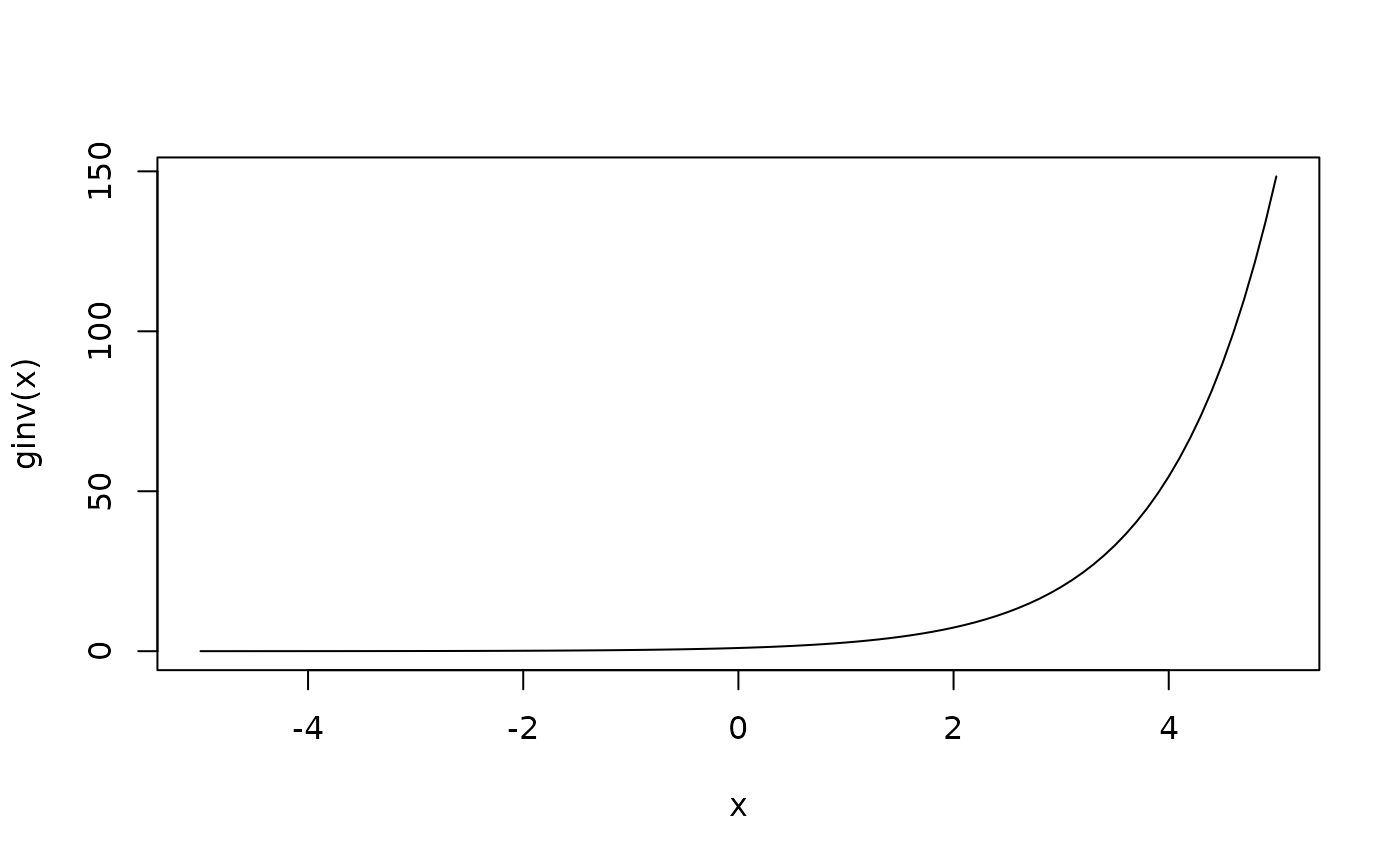
 # Inverse link function
ginv <- log_link()$g_inv
curve(ginv(x), from = -5, to = 5)
# Inverse link function
ginv <- log_link()$g_inv
curve(ginv(x), from = -5, to = 5)